Changelog¶
Explanations of major changes to pixy are listed below.
For up to date info on minor versions and bugfixes, see the current release changelog at https://github.com/ksamuk/pixy/releases
pixy 1.0.0¶
To conincide with the publication of the pixy manuscript, we’re very happy to announce the release pixy version 1.0.0!
This is a major update to pixy, and includes number of major performance increases, new features, simplifications, and many minor fixes. Note that this version contains breaking changes, and old pipelines will need to be updated. We have also validated that the estimates of pi, dxy and fst produced by 1.0.0 are indentical to 0.95.02 (the verison used in the manuscript).
Summary of Major Changes¶
- All calculations are now much faster and natively parallelizable
- Memory usage vastly reduced
- BED and sites file support allows huge flexibility in windows/targeting sites of different classses of genomic elements
- Genotype filtration has been removed
- No change in the core summary statistics (pi, dxy, fst) produced by pixy
- htslib is now a hard dependency, and must be installed separately
- VCFs will need to be compressed with bgzip and indexed with tabix (from htslib) before using pixy
The performance increase and stability of numerical results can be seen in the following plots:
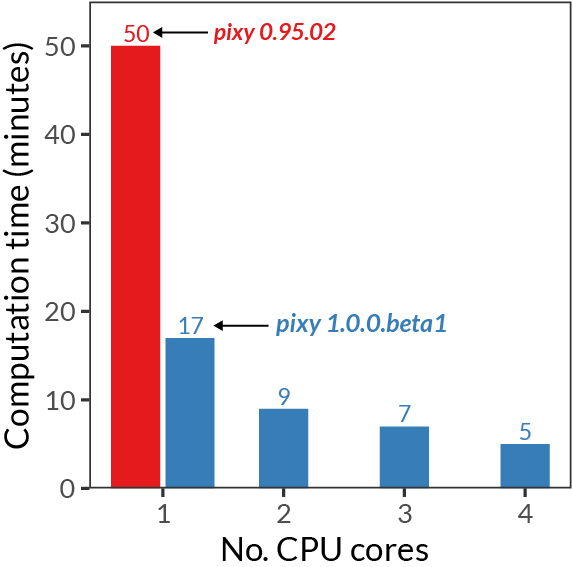
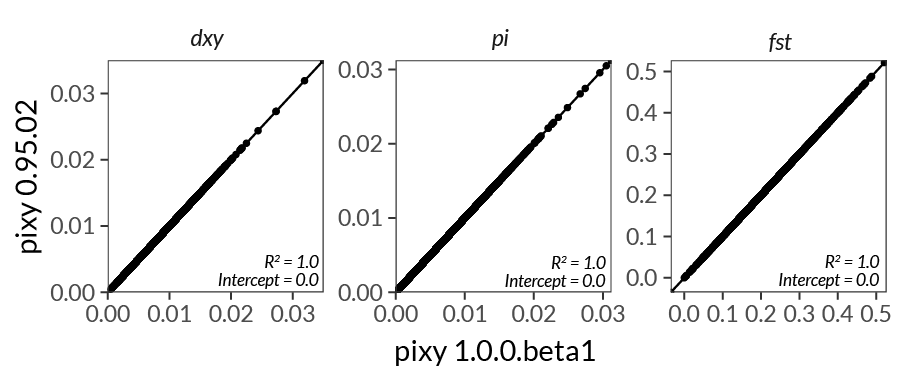
Detailed changelog¶
Major changes¶
- pixy calculations can now be fully parallelized by specifying
--n_cores [number of cores]at the command line.
- Implemented using the multiprocessing module, which is now a hard dependency.
- Supported under both Linux and MacOS (using fork and spawn modes respectively).
- We’ve vectorized many of the core computations performed by pixy using numpy, resulting in significant performance gains.
- The memory usage of pixy is now vastly lower, more intelligently handled, and configurable by the user (via the –chunk_size argument).
- Large windows (e.g. whole chromosomes) are dynamically split into chunks and reassembled after summarization.
- Small windows are assigned to larger chunks to prevent I/O bottlenecks associated with frequently re-reading the source VCF.
New features¶
- Support for BED files specifying windows over which to calculate pi/dxy/fst. These windows can be heterogenous in size.
- This enables precisely matching pixy output with the output of e.g. another program
- Support for a tab-separate ‘sites file’ specifying sites (CHROM, POS) where summary statistics should be exclusively calculated
- This also enables e.g. estimates of pi using only 4-fold degenerate sites, or for only a particular class of genes, etc.
- Basic support for site-level statistics (1bp scale, but note that these are much slower to calculate compared to windowed statistcs)
Removed features¶
- pixy no longer makes use of a Zarr database for storing on-disk intermediate genotype information. We instead now perform random access of the VCF via tabix from htslib as implemented in scikit-allel. As such, htslib is now a hard dependency. We think tabix is a much more flexible system for many datasets, and the performance differences are negliable (and offset by the new performance features in v1.0). VCFs will need to be compressed with bgzip and indexed with tabix before using pixy.
- Other than requiring all variants to be biallelic SNPs, pixy no longer performs any filtration of any kind. We decided that filtration was outside the scope of the functionality we wanted pixy to have. Further, there are many excellent existing tools that perform filtration already and we felt we were “reinventing the wheel”. Further, pre-filtering creates a filtered VCF that can be used for other analyses, which users likely will want to do. We now strongly reccomend that users pre-filter their invariant sites VCFs using VCFtools and/or BCFtools. We provide an example shell script with this functionality (retaining invariant sites as required) as a template for users to edit for their needs.
Minor updates¶
- The pre-calculation checks performed by pixy are now more extensive and systematic.
- The method for calculating the number of valid sites has been slightly ajusted to be more accurate (this was calculated independantly of the pi/dxy/fst statistics).
- We’ve refactored and restructured much of the code, with a focus on increased functionalization. This should make community contributions and future updates much easier.
- To reduce confusion, output prefix and output folder are now separate arguments.
- The documentation for pixy as been extensively updated to reflect the new changes in version 1.0.0.
Other Bugfixes¶
- Total computation time is now properly displayed (issue ref).
- For FST: regions with no variant sites will now have “NA” in the output file, instead of not being represented.